Mass spectrometry (MS) has established itself as an optimized analytical method in the field of drug development due to its high sensitivity and rapid analysis speed. For example, proteins used as drug targets are often present at low concentrations in mixtures, making quantification challenging. However, MS enables concentration analysis. Additionally, high-throughput analysis can be automated and simplified by MS, facilitating the production of accurate and rapid results. These advantages make it suitable for tracking biomarkers in bodily fluids and screening drug-target interactions.
In fact, the term “mass spectrometry” might be considered a misnomer. Rather than measuring the actual mass of substances, MS measures the mass-to-charge ratio (m/z). Therefore, the actual measurement unit is not the unit of mass, but kilograms per Coulomb. As indicated by the measurement unit, MS analyzes ionized molecules, and due to the presence of isotopes, the analysis generally does not reflect the molecular weight as commonly understood.
Fundamentally, MS samples are analyzed in a gaseous ion state. To achieve this, the components of an MS system can be divided into ion sources, analyzers, and detectors.

Based on the flow of the sample within the analytical device, let’s first explore ionization devices. As previously mentioned, samples are analyzed in a gaseous ion state within the MS, which is maintained under a vacuum. Therefore, the process of converting samples into gaseous ions through reactions is crucial. Traditionally, the ionization process has been performed in two stages: volatilization of the sample followed by ionization.
However, this method has limitations because the volatilization stage requires high temperatures, restricting the range of analyzable samples to low-molecular-weight substances. Most biomolecules are high-molecular-weight and highly polar, making volatilization challenging. Advances in technology have allowed for the conversion of temperature-sensitive, non-volatile molecules into gaseous ions. Two major methods that have been adopted for this purpose are electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI).
◎ ESI
ESI, or electrospray ionization, uses a spraying method where the sample dissolved in a solvent pass through a small capillary with an inner diameter of less than 250 µm at atmospheric pressure, generating ions. The small capillary used in this process has a voltage difference of +500 to +4500V compared to the opposing electrode, with the actual voltage varying depending on the needle’s inner diameter and the solvent. Generally, samples with higher boiling points require a higher voltage difference. Once the sample is sprayed, a charged aerosol is created, with nitrogen gas flowing. During the spraying process, the sample is separated into positively or negatively charged solvent and solute (sample), resulting in the sample being isolated from the solvent and existing in an ionized state. The separated sample then flows into the mass spectrometer for analysis.
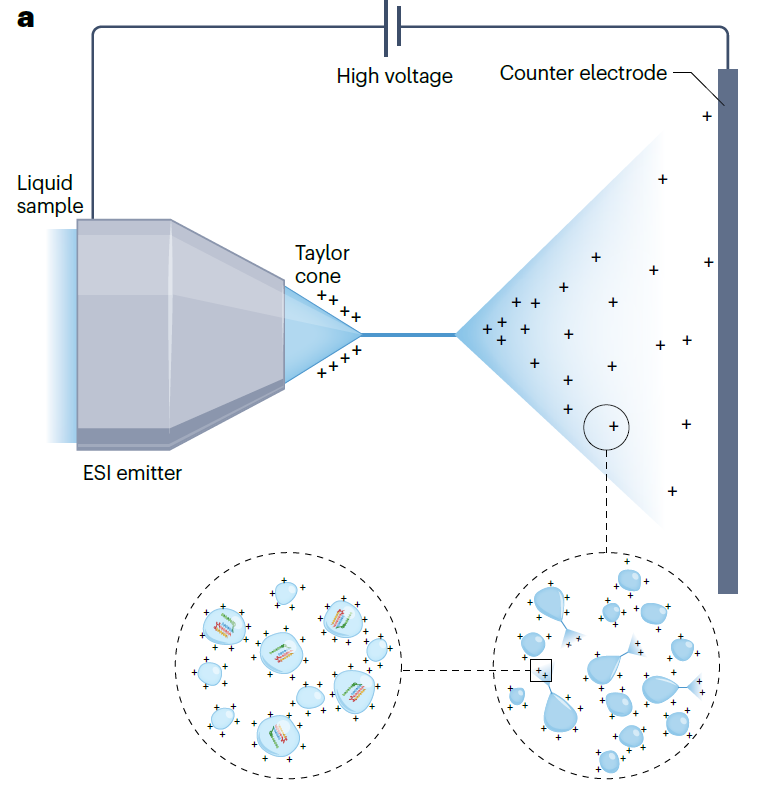
The key principle behind ESI (electrospray ionization) in generating ions from biomolecules is protonation/deprotonation. Biomolecules such as proteins, peptides, and oligonucleotides, which have acidic or basic properties, have multiple sites of protonation/deprotonation. Consequently, depending on the protonation/deprotonation status of each site, multiple peaks appear in the mass spectrum. At first glance, the presence of multiple peaks may seem to complicate molecular weight analysis, but it actually allows for more accurate mass measurements. Additionally, the ability to handle multiple charge states enables the analysis of high molecular weight molecules. For example, cytochrome c has a molecular weight of 12,350 Da, but the addition of 10-20 protons reduces the m/z ratio of the protein to below 2,000, bringing it within the working range of most mass spectrometers.
ESI is known to ionize molecules without size limitations, and it can ionize non-covalent complexes (protein complexes, double-stranded DNA, protein-drug complexes, DNA-drug complexes) without causing damage. However, due to the continuous flow nature of the device, there is a constant consumption of the sample, leading to potential wastage of a certain amount of the sample. Moreover, when dealing with high-concentration samples containing salts above 1 mM, ion generation becomes difficult, resulting in reduced accuracy of the analysis.
A similar technique that can address these drawbacks is atmospheric-pressure chemical ionization (APCI). Unlike ESI, which uses voltage for ionization, APCI uses nitrogen and heat, and the ionization of the sample occurs through a chain reaction initiated by a corona discharge. This method is less sensitive to salts and can easily ionize weakly polar molecules. However, due to the use of heat, APCI is not suitable for temperature-sensitive substances and requires high-purity samples. Therefore, APCI can be used as a complementary technology to ESI.
◎ MALDI
Unlike ESI, where sample ions are continuously introduced, ions in MALDI (matrix-assisted laser desorption/ionization) are generated by a pulsed laser. In MALDI, the sample is mixed with a solid matrix that absorbs the laser wavelength and is then crystallized onto a probe. When the laser irradiates the probe, the matrix molecules absorb the energy and facilitate the desorption of the sample, which becomes protonated, resulting in the formation of gas-phase ions. These ions then similarly enter the mass analyzer to ESI.
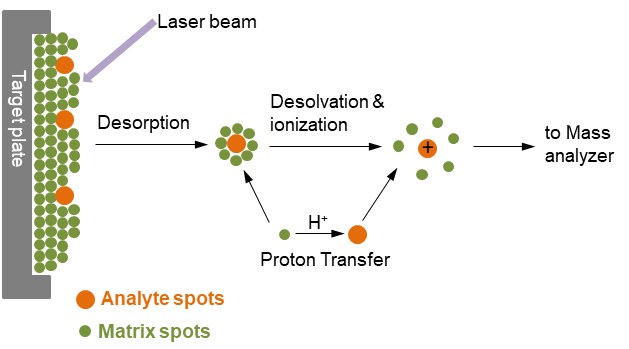
Although singly protonated samples are typically generated, the precise mechanism of ion formation in MALDI is not yet fully understood. A key difference from ESI is that the laser irradiates in a pulsed pattern, creating distinct ions separately, which minimizes sample wastage. Additionally, because MALDI generally produces singly charged ions, it is advantageous for rapid analysis when coupled with a TOF (Time of Flight) mass spectrometer. MALDI also exhibits higher resistance to salts and solvents, enabling the direct ionization of proteins from cell lysates.
However, there are some limitations. MALDI’s pulsed ionization mechanism is not compatible with all types of mass spectrometers. Moreover, the matrices used in MALDI can generate significant chemical noise around 500 Da, making it challenging to analyze low-molecular-weight compounds. To overcome this limitation, techniques like Desorption/Ionization on Silicon (DIOS) have been developed, reducing noise and allowing for the analysis of low-molecular-weight substances.
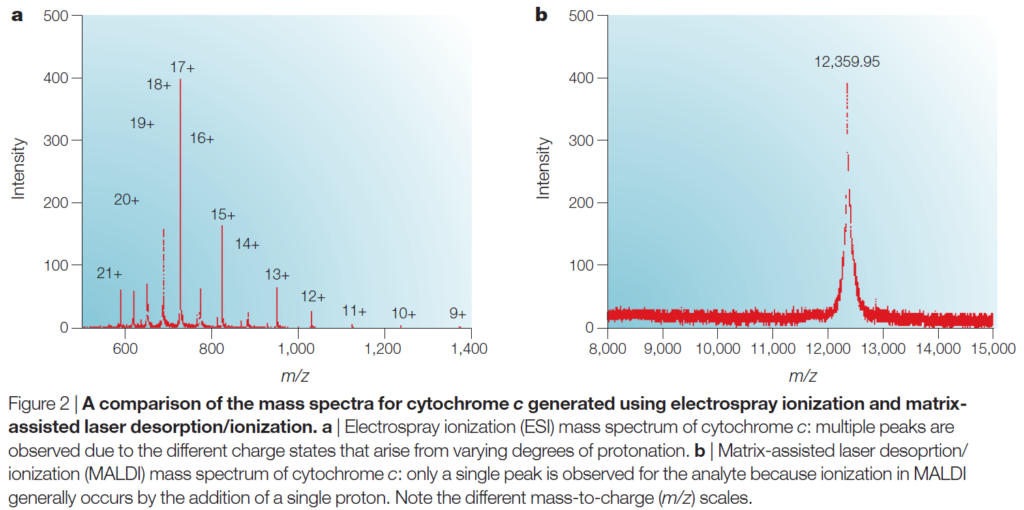
The following two graphs depict cytochrome C ionized using two different methods: ESI (left) and MALDI (right). When utilizing ESI, multiple peaks appear on the graph due to the presence of several protonation/deprotonation sites within the protein molecule. In contrast, with MALDI, ionization occurs with only one protonation, resulting in the observation of a single peak on the graph.
Thus, the ionization method can be selected based on the characteristics of the sample and the purpose of the analysis to conduct mass spectrometry. In the next section, we will explore the different types of mass analyzers and their principles for measuring the mass-to-charge ratio (m/z) of ions.
참고문헌 : Glish, G., Vachet, R. The basics of mass spectrometry in the twenty-first century. Nat Rev Drug Discov 2, 140–150 (2003). https://doi.org/10.1038/nrd1011
Figure 2: Prabhu, G.R.D., Williams, E.R., Wilm, M. et al. Mass spectrometry using electrospray ionization. Nat Rev Methods Primers 3, 23 (2023). https://doi.org/10.1038/s43586-023-00203-4
Figure 3: https://www.creative-proteomics.com/technology/maldi-tof-mass-spectrometry.htm
